Carlo Brogna, Simone Cristoni, Giuliano Marino, Luigi Montano, Valentina Viduto, Mark Fabrowski, Gennaro Lettieri, Marina Piscopo, 31 August 2023
https://doi.org/10.1002/prca.202300048
Abstract
Purpose
The SARS-CoV-2 pandemic prompted the development and use of next-generation vaccines. Among these, mRNA-based vaccines consist of injectable solutions of mRNA encoding for a recombinant Spike, which is distinguishable from the wild-type protein due to specific amino acid variations introduced to maintain the protein in a prefused state. This work presents a proteomic approach to reveal the presence of recombinant Spike protein in vaccinated subjects regardless of antibody titer.
Experimental design
Mass spectrometry examination of biological samples was used to detect the presence of specific fragments of recombinant Spike protein in subjects who received mRNA-based vaccines.
Results
The specific PP-Spike fragment was found in 50% of the biological samples analyzed, and its presence was independent of the SARS-CoV-2 IgG antibody titer. The minimum and maximum time at which PP-Spike was detected after vaccination was 69 and 187 days, respectively.
Conclusions and clinical relevance
The presented method allows to evaluate the half-life of the Spike protein molecule “PP” and to consider the risks or benefits in continuing to administer additional booster doses of the SARS-CoV-2 mRNA vaccine. This approach is of valuable support to complement antibody level monitoring and represents the first proteomic detection of recombinant Spike in vaccinated subjects.
1 INTRODUCTION
Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2) is the causative coronavirus of COVID-19 respiratory disease (coronavirus disease 2019), responsible for the ongoing pandemic that holds the entire world in check. Researchers around the world during these pandemic years have studied this virus, trying to understand its mechanism of action [1–11]. The RNA genome of SARS-CoV-2 consists of about 30,000 nucleotides and contains 11 major coding genes. From a structural point of view, SARS-CoV-2 is characterized by a large number of glycosylated Spike (S) proteins covering its surface and facilitating binding to the angiotensin-converting enzyme 2 (ACE2) receptor of the host cell, mediating viral cell entry [12]. The Spike protein (S) is one of the four major proteins of SARS-CoV-2 [13]. It enables recognition of the host cell receptor and subsequent entry into the host cell. It consists of the distal S1 subunit, which is useful for recognition, and the proximal S2 subunit, which is essential for fusion with the host cell membrane [13]. During the development of the two most widely used mRNA-based vaccines, Pfizer-BioNTech (BNT162b2- Comirnaty) and Moderna (mRNA-1273), all uridine nitrogen bases were replaced with methyl pseudouridine (m1Ψ)—a less immunogenic [14] but more stable nitrogen base [15]. At the same time, mutations were made within the 4284 nucleotides constituting the Spike protein, at positions K986P and V987P to make the protein produced after ribosomal reading stable prefusion form to stimulate increased production of human antibodies (Figure 1A) [15]. The S protein of SARS-CoV-2 is highly conserved among all human coronaviruses (HCoV) and is involved in receptor recognition, viral attachment, and entry into the host cells. For this reason, it represents, for a part of scientists, one of the most important targets for the development of vaccines and therapeutic approaches against COVID-19. Among the COVID-19 vaccines developed and tested, those that have shown the most promising results in preventing COVID-19 infection are a novel class of vaccine products composed of messenger ribonucleic acid (mRNA) filaments encapsulated in lipid nanoparticles (LNPs). Two of them have received “emergency use authorization” (from the Federal and Drug Administration) and “conditional approval” (from the European Medicines Agency). Both consist of a recombinant mRNA to be inoculated as a vaccine, which encodes for a recombinant SARS-CoV-2 Spike protein. Although the mRNAs are different, both encode for the same recombinant Spike protein (here called PP-Spike). This differs from the natural one (wt-Spike, Figure 1B) by a double amino acid change at position 986 and 987 (K986P and V987P, i.e., the amino acids lysine and valine are both replaced by two proline amino acids) [16, 17], in order to stabilize the Spike conformation in an inactive prefusion state (Figure 1A). The introduced double amino acid variation abolishes a tryptic digestion site. As a result, it is possible to distinguish, by tryptic digestion [18], followed by mass spectrometry analysis [19–22], synthetic Spike proteins originated from the translation of the mRNA vaccines from natural Spike circulating in biological fluids. Here we present a methodological approach that can specifically detect the presence of PP-Spike in biological fluids of human and animal organisms, such as blood, urine, saliva, and bronchoalveolar lavage fluids.
Significance of the study
Although the COVID-19 pandemic brought the whole world to its knees, it also allowed many scientists to develop ideas and solutions against viruses. These include mRNA vaccines, which, due to their versatility in production, may represent a new standard of vaccination. However, it is the duty of the scientist not to neglect controls. Herein lies the importance of monitoring vaccine-induced Spike protein “PP” after a time period after vaccination in human biological samples. The presented method allows to assess the half-life of the Spike “PP” protein molecule and to consider the risks or benefits in continuing further booster doses of the SARS-CoV-2 mRNA vaccine.
FIGURE 1

Schematic diagram of PP Spike protein production. (A) Formation of Spike PP protein after injection of vaccine mRNA. (B) Example of wild-type, unmodified Spike protein. (C) Extraction and detection scheme (more details in the text). (D) quantification of the antibody titer of the 20 vaccinated cases. (E) Time from the day of vaccination completion to the discovery of vaccine mRNA-induced Spike protein “PP”.
The study group, from southern Italy, was 40 subjects, 20 were vaccinated with the full cycle of mRNA vaccine as of April 2022, being part of the health sector, and 20 were unvaccinated with negativity for COVID-19 to nasopharyngeal test and with no titer of any antibodies. Other 20 unvaccinated persons were added that were positive for COVID-19. The specific PP-Spike fragment was found in 50% of the biological samples analyzed (Figures 1C–E and 2). This presence was independent of the SARS-CoV-2 IgG antibody titer. The antibody titers had a geometric mean of 629.86 BAU/mL (Figure 1E). The minimum time PP-Spike was detected was 69 days after vaccination, while the maximum time was 187 days. All controls (samples from unvaccinated individuals) were negative. The control group (20 unvaccinated people) was also tested after contracting COVID-19 and was negative for PP-spike.
FIGURE 2
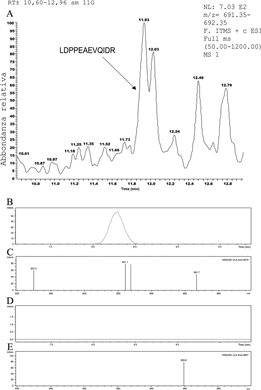
Molecular ion of the sequence LDPPEAEVQIDR- Spike “PP.” (A) Shows the m/z (molecular ion) ratio of the SEQ ID NO:1 detection. Shows an example of the spectrum of the peptide zone marked by the enzymatic failure to cut the “PP” amino acids of the “Spike” protein induced by mRNA vaccines. (B-E) Extraction mass chromatograms of (B) vaccinated subjects and (D) control samples were obtained by fragmenting the [M+3H]3+ion at m/z 461 and monitoring the singly charged signal at m/z 851. Also shown are the tandem mass spectra obtained by analyzing (C) the vaccinated subject and (E) the control subject. Five microliters of digested samples were injected, and the eluent flow rate was 250 microliter/min.
Some studies [23] have observed the presence of the vaccine Spike protein immediately after injection.
According to the authors [15] and in general, the vaccine messenger RNA nanoparticle molecules should be picked up by the immune cells in the lymph nodes after injection into the muscle. Recently, other authors have isolated vaccine messenger RNA sequences from peripheral plasma after 28 days after injection [24]. The question of whether or not the vaccine RNA can be integrated into the lymphocyte or other body cells is much debated. Nonetheless, the observation of the protein produced, as presented in this manuscript, goes beyond the purely cognitive aspect and defines a method to verify not only the persistence of the vaccine RNA but the quantification of the product, that is, the protein that is supposed to induce antibody production, in order to verify the correct half-life and a possible need to update the vaccine status. Using mass spectrometry examination of biological samples, we detected the presence of specific fragments of recombinant Spike protein in about 50% of subjects who received mRNA-based vaccines. In some cases, we found the PP-Spike marker in vaccinated individuals more than 30 days after the vaccine, indicating that it is possible to detect vaccine “Spike” protein even sometime after vaccination and in any organic tissue (data in preparation). Based on the results obtained, hypotheses can be made for possible molecular mechanisms of persistence of “Spike PP.” In particular, three hypotheses are possible and are shown in Figure 3.
- It is possible that the mRNA may be integrated or re-transcribed in some cells.
- It is possible that pseudo-uridines at a particular sequence position, as described in the article, induce the formation of a spike protein that is always constitutively active. But it seems very remote as a hypothesis.
- It is possible that the mRNA-containing nanoparticle will be picked up by bacteria normally present at the basal level in the blood. In fact, the existence of blood microbiota in clinically healthy individuals was proven during the last 50 years. Indeed, indirect evidence by radiometric analyses suggested the existence of living microbial forms in erythrocytes [25]. In addition, the observation of the PP-Spike marker in individuals vaccinated more than 30 days after the vaccine in about 50% of subjects could also be explained by the wide biodiversity of eukaryotic and prokaryotic microbiota identified in blood by next-generation sequencing technologies [25].
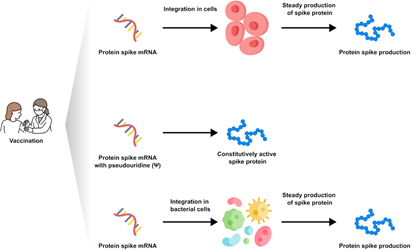
FIGURE 3
Possible molecular mechanisms of persistence of “Spike PP”.
In conclusion, the possibility of detecting the presence of specific fragments of recombinant Spike protein opens new scenarios for monitoring the presence and half-life of vaccine Spike protein in vaccinated individuals. The hypotheses advanced need more and larger studies. At present, these initial observations are limited only to assessing the presence of the vaccine protein with the aim of wanting to provide aid in the individual’s decision to administer booster doses or temporize.
2 ASSOCIATED DATA
Statistical analysis: t-test with © 2023 GraphPad Software was performed between the two groups, vaccinated, and not vaccinated for the presence of the peptide of spike called PP. “P-value and statistical significance: The two-tailed P value is equal to 0.0010. Confidence interval: The mean of group one minus group two is equal to 0.40, 95% confidence interval of this difference: 0.17–0.63. Intermediate values used in calculations: t = 35,590, df = 38, standard error of the difference = 0.112. The difference in titer antibody between the two groups is not significant.
3 EXPERIMENTAL PROCEDURES
In accordance with applicable territorial regulations, case reports, case series, and observational retrospective studies do not require ethics committee approval. Informed consent expressed by all participants, South of Italy, was obtained. Twenty human biological samples were collected from vaccinated subjects with express, free, and informed consent for collection and use. The geometric mean of their antibodies′ titer versus spike protein was 629,86 BAU/mL after 60 days from vaccination. In addition, 20 human biological samples were collected from unvaccinated subjects (control group, negative) with express, free, and informed consent for collection and use. Candidates had not previously undergone COVID-19 and were negative for molecular testing by nasopharyngeal swab, and the antibodies titer was negative. Other 20 human biological samples from unvaccinated patients, who were ill with COVID-19, were obtained (geometric mean of antibodies titer vs. spike protein was 187,89 BAU/mL later 105 days from disease).
3.1 Pre-analytic
Preclinical dry blood sample collection was performed in compliance with the scientific literature: the Dry Blood Spot (DBS) technique is common for the collection of small amounts of blood, especially from newborns for screening application. This method was first published by Robert Guthrie in the early 1960s and is based on sampling a small blood spot onto a filter paper instead of drawing a few milliliters of blood into a vial. With the DBS technique, sampling is easy and fast without the need for trained professionals. However, the small amounts of blood drawn (∼20–40 µL) make this approach more analytically challenging in terms of sample preparation and sensitivity compared with traditional blood sampling [26, 27]. Blood drops were collected in sterile mode. In order to preserve the protein structure, the dry blood filter was maintained at −20°C conditions until the processing [28].
3.2 Rationale
Trypsin [18] is an enzyme belonging to the class of hydrolases capable of reducing proteins to smaller polypeptides by proteolytic cuts with specificity for arginine (R) and lysine (K). Synthetic Spike protein and natural Spike protein can thus be distinguished as they produce different tryptic digestion products:
- – When digested by trypsin, the PP-Spike encoded by vaccine mRNA produces an LDPPEAEVQIDR fragment (PP-Spike marker) (Figure 2C).
- – The wild-type SAR-CoV-2 protein, when digested by trypsin, produces two smaller fragments, namely LDK + VEAEVQIDR.
More than 6,600,000 SARS-CoV-2 genomes have been sequenced, and it appears that none of them have K986P and V987P mutations, including the Omicron variant [29].
Buffers used: Double distilled water (VWR); ammonium bicarbonate (NH4HCO3) (Sigma Aldrich). Reagents: Trypsin from Promega.
Reagent preparation: Trypsin solution 25 ng/µL.
Preparation procedure: Resuspend, in a vial, 20 µg of solid trypsin in 800 µL of 50 mM NH4HCO3 solution. Vortex the vial until the trypsin is completely dissolved.
The standard peptide corresponding to the fragment LDPPEAEVQIDR (Sigma Aldrich, UK) has been diluted to a concentration of 10 ng/mL in bi-distilled water. Ten microliters were injected into the LC-MS apparatus for control purposes. The peptide detected in plasma was characterized on the basis of EU regulations [30].
3.3 Enzymatic digestion
The enzymatic digestion procedure of blood drops is performed under a fume hood in order to minimize operator exposure to any form of chemical biohazard. The following method was used for each blood sample. Place 2 µL of capillary blood into a labeled Eppendorf tube; add 40 µL of trypsin (Promega, Italy) solubilized in a 50 mmoL NH4HCO3 solution to the blood sample; vortex for 30 s; test the pH, which must be between 7 and 8; transfer the Eppendorf tube to the thermoblock; incubate for 2 days at 37°C; add 40 µL of NH4HCO3 50 mmoL. Collect 40 µL of the supernatant obtained after centrifugation at 13000 G for 10 min and transfer to an Eppendorf tube. Add 2 µL of pure formic acid and transfer the solution to an injection vial. The vial is inserted into the autosampler coupled to the mass spectrometer, and 5 µL is injected into the chromatographic column.
3.4 LC–SACI–MS instrumentation
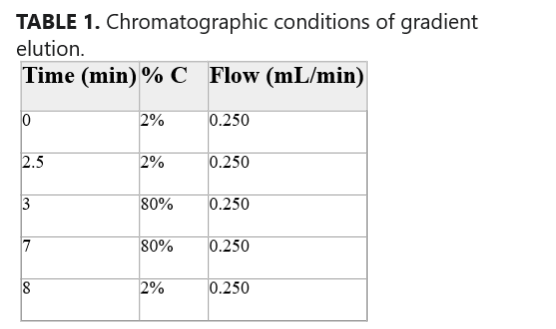
The analysis was performed with a Surveyor MS HPLC (Thermofisher, USA). The column used was a Kinetex 50 × 4.6 mm 2.6 µm. The analysis was performed using a two-phase gradient: Phase A (H2O + 0.2 % formic acid (HCOOH)) and Phase C (CH3CN) (Table 1). The chromatographic gradient used is shown in Table 1. The volume of the sample injected is 5 µL. The ionization source used is a SACI-ESI. A surface potential of 0 V, a nebulizer gas pressure of 75 PSI, and a dry gas flow of 1.0 L/min were used. The dry gas temperature is at 320°C [19–22].
TABLE 1. Chromatographic conditions of gradient elution.
| Time (min) | % C | Flow (mL/min) |
| 0 | 2% | 0.250 |
| 2.5 | 2% | 0.250 |
| 3 | 80% | 0.250 |
| 7 | 80% | 0.250 |
| 8 | 2% | 0.250 |
- Note: The percentage of eluent phase C (CH3CN) is reported along with the eluent flow rate at different gradient elution times.
Purpose
The SARS-CoV-2 pandemic prompted the development and use of next-generation vaccines. Among these, mRNA-based vaccines consist of injectable solutions of mRNA encoding for a recombinant Spike, which is distinguishable from the wild-type protein due to specific amino acid variations introduced to maintain the protein in a prefused state. This work presents a proteomic approach to reveal the presence of recombinant Spike protein in vaccinated subjects regardless of antibody titer.
Experimental design
Mass spectrometry examination of biological samples was used to detect the presence of specific fragments of recombinant Spike protein in subjects who received mRNA-based vaccines.
Results
The specific PP-Spike fragment was found in 50% of the biological samples analyzed, and its presence was independent of the SARS-CoV-2 IgG antibody titer. The minimum and maximum time at which PP-Spike was detected after vaccination was 69 and 187 days, respectively.
Conclusions and clinical relevance
The presented method allows to evaluate the half-life of the Spike protein molecule “PP” and to consider the risks or benefits in continuing to administer additional booster doses of the SARS-CoV-2 mRNA vaccine. This approach is of valuable support to complement antibody level monitoring and represents the first proteomic detection of recombinant Spike in vaccinated subjects.
REFERENCES
- 1Brogna, B., Brogna, C., Petrillo, M., Conte, A. M., Benincasa, G., Montano, L., & Piscopo, M. (2021). SARS-CoV-2 detection in fecal sample from a patient with typical findings of COVID-19 pneumonia on CT but negative to multiple SARS-CoV-2 RT-PCR tests on oropharyngeal and nasopharyngeal swab samples. Medicina, 57(3), 3. https://doi.org/10.3390/medicina57030290
- 2Brogna, C., Brogna, B., Bisaccia, D. R., Giuliano, M., Montano, L., Cristoni, S., Petrillo, M., & Piscopo, M. (2022). SARS-CoV-2: Reinfection after 18 months of a previous case with multiple negative nasopharyngeal swab tests and positive fecal molecular test. Medicina, 58(5), 5. https://doi.org/10.3390/medicina58050642
- 3Brogna, C., Brogna, B., Bisaccia, D. R., Lauritano, F., Marino, G., Montano, L., Cristoni, S., Prisco, M., & Piscopo, M. (2022). Could SARS-CoV-2 have bacteriophage behavior or induce the activity of other bacteriophages? Vaccines, 10(5), 5. https://doi.org/10.3390/vaccines10050708
- 4Brogna, C., Costanzo, V., Brogna, B., Bisaccia, D. R., Brogna, G., Giuliano, M., Montano, L., Viduto, V., Cristoni, S., Fabrowski, M., & Piscopo, M. (2023). Analysis of bacteriophage behavior of a human RNA virus, SARS-CoV-2, through the integrated approach of immunofluorescence microscopy, proteomics and D-amino acid quantification. International Journal of Molecular Sciences, 24(4), 4. https://doi.org/10.3390/ijms24043929
- 5Brogna, C., Cristoni, S., Brogna, B., Bisaccia, D. R., Marino, G., Viduto, V., Montano, L., & Piscopo, M. (2023). Toxin-like peptides from the bacterial cultures derived from gut microbiome infected by SARS-CoV-2—new data for a possible role in the long COVID pattern. Biomedicines, 11(1), 1. https://doi.org/10.3390/biomedicines11010087
- 6Gu, W., Gan, H., Ma, Y., Xu, L., Cheng, Z. J., Li, B., Zhang, X., Jiang, W., Sun, J., Sun, B., & Hao, C. (2022). The molecular mechanism of SARS-CoV-2 evading host antiviral innate immunity. Virology Journal, 19(1), 49. https://doi.org/10.1186/s12985-022-01783-5
- 7Gusev, E., Sarapultsev, A., Solomatina, L., & Chereshnev, V. (2022). SARS-CoV-2-specific immune response and the pathogenesis of COVID-19. International Journal of Molecular Sciences, 23(3), 1716. https://doi.org/10.3390/ijms23031716
- 8Patel, R., Kaki, M., Potluri, V. S., Kahar, P., & Khanna, D. (2022). A comprehensive review of SARS-CoV-2 vaccines: Pfizer, Moderna & Johnson & Johnson. Human Vaccines & Immunotherapeutics, 18(1), 2002083. https://doi.org/10.1080/21645515.2021.2002083
- 9Săndulescu, O., Apostolescu, C. G., Preoțescu, L. L., Streinu-Cercel, A., & Săndulescu, M. (2023). Therapeutic developments for SARS-CoV-2 infection—molecular mechanisms of action of antivirals and strategies for mitigating resistance in emerging variants in clinical practice. Frontiers in Microbiology, 14, 1132501. https://www.frontiersin.org/articles/10.3389/fmicb.2023.1132501
- 10Sasidharan, S., Sarkar, N., & Saudagar, P. (2023). Discovery of compounds inhibiting SARS-COV-2 multi-targets. Journal of Biomolecular Structure and Dynamics, 41(6), 2602–2617. https://doi.org/10.1080/07391102.2021.2025149
- 11Silva Andrade, B., Siqueira, S., de Assis Soares, W. R., de Souza Rangel, F., Santos, N. O., dos Santos Freitas, A., Ribeiro da Silveira, P., Tiwari, S., Alzahrani, K. J., Góes-Neto, A., Azevedo, V., Ghosh, P., & Barh, D. (2021). Long-COVID and post-COVID health complications: An up-to-date review on clinical conditions and their possible molecular mechanisms. Viruses, 13(4), 4. https://doi.org/10.3390/v13040700
- 12Letko, M., Marzi, A., & Munster, V. (2020). Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature Microbiology, 5(4), 562–569. https://doi.org/10.1038/s41564-020-0688-y
- 13Dai, L., & Gao, G. F. (2021). Viral targets for vaccines against COVID-19. Nature Reviews Immunology, 21(2), 2. https://doi.org/10.1038/s41577-020-00480-0View
- 14Andries, O., Mc Cafferty, S., De Smedt, S. C., Weiss, R., Sanders, N. N., & Kitada, T. (2015). N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. Journal of Controlled Release: Official Journal of the Controlled Release Society, 217, 337–344. https://doi.org/10.1016/j.jconrel.2015.08.051View
- 15Nance, K. D., & Meier, J. L. (2021). Modifications in an emergency: The role of N1-methylpseudouridine in COVID-19 vaccines. ACS Central Sciences, 7(5), 748–756. https://doi.org/10.1021/acscentsci.1c00197View
- 16Corbett, K. S., Edwards, D. K., Leist, S. R., Abiona, O. M., Boyoglu-Barnum, S., Gillespie, R. A., Himansu, S., Schäfer, A., Ziwawo, C. T., DiPiazza, A. T., Dinnon, K. H., Elbashir, S. M., Shaw, C. A., Woods, A., Fritch, E. J., Martinez, D. R., Bock, K. W., Minai, M., Nagata, B. M., & Graham, B. S. (2020). SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature, 586(7830), 567–571. https://doi.org/10.1038/s41586-020-2622-0
- 17Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., Graham, B. S., & McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science, 367(6483), 1260–1263. https://doi.org/10.1126/science.abb2507
- 18Olsen, J. V., Ong, S.-E., & Mann, M. (2004). Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Molecular & Cellular Proteomics: MCP, 3(6), 608–614. https://doi.org/10.1074/mcp.T400003-MCP200
- 19Cristoni, S., & Bernardi, L. R. (2004). Bioinformatics in mass spectrometry data analysis for proteomics studies. Expert Review of Proteomics, 1(4), 469–483. https://doi.org/10.1586/14789450.1.4.469
- 20Cristoni, S., Bernardi, L. R., Biunno, I., Tubaro, M., & Guidugli, F. (2003). Surface-activated no-discharge atmospheric pressure chemical ionization. Rapid Communications in Mass Spectrometry, 17(17), 1973–1981. https://doi.org/10.1002/rcm.1141
- 21Lundgren, D. H., Hwang, S.-I., Wu, L., & Han, D. K. (2010). Role of spectral counting in quantitative proteomics. Expert Review of Proteomics, 7(1), 39–53. https://doi.org/10.1586/epr.09.69
- 22Okada, P., Buathong, R., Phuygun, S., Thanadachakul, T., Parnmen, S., Wongboot, W., Waicharoen, S., Wacharapluesadee, S., Uttayamakul, S., Vachiraphan, A., Chittaganpitch, M., Mekha, N., Janejai, N., Iamsirithaworn, S., Lee, R. T., & Maurer-Stroh, S. (2020). Early transmission patterns of coronavirus disease 2019 (COVID-19) in travellers from Wuhan to Thailand, January 2020. Eurosurveillance, 25(8), 2000097. https://doi.org/10.2807/1560-7917.ES.2020.25.8.2000097
- 23Cognetti, J. S., & Miller, B. L. (2021). Monitoring serum spike protein with disposable photonic biosensors following SARS-CoV-2 vaccination. Sensors, 21(17), 17. https://doi.org/10.3390/s21175857
- 24Castruita, J. A. S., Schneider, U. V., Mollerup, S., Leineweber, T. D., Weis, N., Bukh, J., Pedersen, M. S., & Westh, H. (2023). SARS-CoV-2 spike mRNA vaccine sequences circulate in blood up to 28 days after COVID-19 vaccination. Apmis, 131(3), 128–132. https://doi.org/10.1111/apm.13294
- 25Tsafarova, B., Hodzhev, Y., Yordanov, G., Tolchkov, V., Kalfin, R., & Panaiotov, S. (2023). Morphology of blood microbiota in healthy individuals assessed by light and electron microscopy. Frontiers in Cellular and Infection Microbiology, 12, 1091341. https://www.frontiersin.org/articles/10.3389/fcimb.2022.1091341
- 26Yishai Aviram, L., Magen, M., Chapman, S., Neufeld Cohen, A., Lazar, S., & Dagan, S. (2018). Dry Blood Spot sample collection for post-exposure monitoring of chemical warfare agents—in vivo determination of phosphonic acids using LC-MS/MS. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences, 1093-1094, 60–65. https://doi.org/10.1016/j.jchromb.2018.06.035
- 27 Medical Advisory Secretariat. (2003). Neonatal screening of inborn errors of metabolism using tandem mass spectrometry: An evidence-based analysis. Ontario Health Technology Assessment Series, 3(3), 1–36.
- 28Di Girolamo, F., Alessandroni, J., Somma, P., & Guadagni, F. (2010). Pre-analytical operating procedures for serum low molecular weight protein profiling. Journal of Proteomics, 73(3), 667–677. https://doi.org/10.1016/j.jprot.2009.09.006
- 29Miller, N. L., Clark, T., Raman, R., & Sasisekharan, R. (2021). Insights on the mutational landscape of the SARS-CoV-2 Omicron variant. Cell Reports Medicine, 3(2), 100527. https://doi.org/10.1016/j.xcrm.2022.1005279
- 30Cristoni, S., Dusi, G., Brambilla, P., Albini, A., Conti, M., Brambilla, M., Bruno, A., Di Gaudio, F., Ferlin, L., Tazzari, V., Mengozzi, S., Barera, S., Sialer, C., Trenti, T., Cantu, M., Rossi Bernardi, L., & Noonan, D. M. (2017). SANIST: Optimization of a technology for compound identification based on the European Union directive with applications in forensic, pharmaceutical and food analyses. Journal of Mass Spectrometry: JMS, 52(1), 16–21. https://doi.org/10.1002/jms.3895
