Authors: Magda Gioia,a,⁎ Chiara Ciaccio,a,⁎ Paolo Calligari,b Giovanna De Simone,c Diego Sbardella,d Grazia Tundo,dGiovanni Francesco Fasciglione,a Alessandra Di Masi,cD onato Di Pierro,a Alessio Bocedi,b Paolo Ascenzi,c,e and Massimo Colettaa,⁎ Biochem Pharmacol. 2020 Dec; 182: 114225.Published online 2020 Sep doi: 10.1016/j.bcp.2020.114225PMCID: PMC7501082PMID: 32956643
Abstract
In the Fall of 2019 a sudden and dramatic outbreak of a pulmonary disease (Coronavirus Disease COVID-19), due to a new Coronavirus strain (i.e., SARS-CoV-2), emerged in the continental Chinese area of Wuhan and quickly diffused throughout the world, causing up to now several hundreds of thousand deaths.
As for common viral infections, the crucial event for the viral life cycle is the entry of genetic material inside the host cell, realized by the spike protein of the virus through its binding to host receptors and its activation by host proteases; this is followed by translation of the viral RNA into a polyprotein, exploiting the host cell machinery. The production of individual mature viral proteins is pivotal for replication and release of new virions.
Several proteolytic enzymes either of the host and of the virus act in a concerted fashion to regulate and coordinate specific steps of the viral replication and assembly, such as (i) the entry of the virus, (ii) the maturation of the polyprotein and (iii) the assembly of the secreted virions for further diffusion. Therefore, proteases involved in these three steps are important targets, envisaging that molecules which interfere with their activity are promising therapeutic compounds.
In this review, we will survey what is known up to now on the role of specific proteolytic enzymes in these three steps and of most promising compounds designed to impair this vicious cycle.Go to:
1. Introduction
Over the past twenty years, β-Coronaviruses (CoV)s have caused three epidemics/pandemics, namely SARS-CoV in 2002, MERS-CoV in 2013 and SARS-CoV-2 in 2019, which have been associated with acute severe respiratory illnesses. As of September 8, SARS-CoV-2 virus, responsible for the global COVID-19 pandemic, has been causing more than 27 millions of contagions and around 900.000 deaths worldwide (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports). The scientific community has come together to understand the mechanisms underlying the infection and the virulence of SARS-CoV-2 virus, as well as all symptoms and risk factors for subsequent mortality. Currently, there is neither antiviral nor vaccine for the severe acute respiratory syndrome induced by SARS-CoV-2. Its alarming spread and its severity across most countries elicited an effort for the elucidation of the pathogenetic features of this novel viral infection in search for effective therapeutic approaches [1].
Little is known about the pathobiology of SARS-CoV-2, even though the availability of the virus genome sequence (GenBank ID: MN908947.3) has demonstrated crucial similarities between SARS-CoV-2 and other members of the same viral order (i.e., Nidovirales) [2]. Hence, to rapidly gain insights on molecular mechanism of SARS-CoV-2 it is worth exploiting what we have learned from several medicinal chemistry studies on viral spreading to help us in finding promising targets for the development of anti-viral strategies for SARS-CoV-2.
1.1. Host and viral proteases involved in viral life-cycle
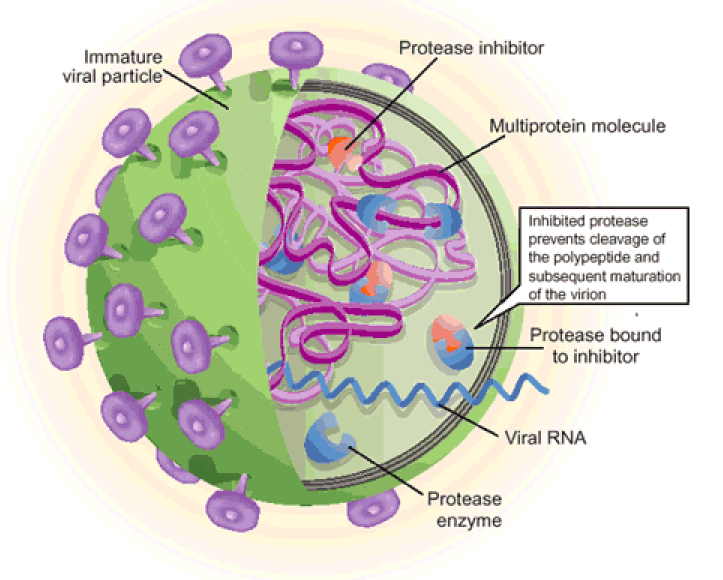
As for common viral infections, the crucial event for the viral life cycle is the entry of genetic material inside the host cell for replication and release of new virions. During its life-cycle, SARS-CoV-2 is internalized in the host cell where the viral RNA is translated, exploiting the host cell machinery and giving rise to virus-encoded proteins of different open reading frames (ORF)s. The ORF1, which encompasses about 75% of the viral genome, is translated into two viral replicase polyproteins (i.e., pp1a and pp1ab) (Fig. 1 ). Sixteen mature non-structural proteins (nsp) arise from further processing of these two pps, which are autocatalytically processed by two proteases (also auto-processed), namely (a) the papain-like protease (PLpro), which cleaves the first two non-structural proteins (nsp1 and nsp2) at the N-terminal region of the polyprotein, and (b) the main protease (Mpro, also known as a chymotrypsin-like cysteine protease, 3CLpro), which recognizes cleavage sites at the C-terminus and brings to the production of about 11 individual mature non-structural proteins [5], [3], [4]. The remaining ORFs encode accessory and structural proteins, like spike surface glycoprotein (S), small envelope protein (E), matrix protein (M), nucleocapsid protein (N) (see Fig. 1).

SARS-CoV-2 polyproteins encoded by ORF1a and ORF1ab. Schematic representation of the open reading frames 1a and 1ab, which encode for polyproteins pp1a and pp1ab. Proteins composing each polyprotein are shown: (ns) indicates non-structural proteins; RNA dependent RNA polymerase and Helicase are indicated by (RdRp) and (Hel), respectively. Proteolytic sites cleaved by PLpro and Mpro are reported in yellow and green arrows, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Since proteolytic enzymes are the major actors of the various events described in this review and although knowledge about their role is continuously expanding, it may be worth recalling that they can be roughly classified into seven broad groups (from the type of aminoacid involved as proton donor for the activation of the peptide bond to be cleaved), namely (a) serine protease, (b) cysteine protease, (c) threonine protease, (d) aspartic protease, (e) glutamic protease, (f) metalloproteases (usually employing Zn++), and (g) asparagine peptide lyases [6], [7]. Within each group, a further differentiation can be applied according to whether the peptide bond cleaved by the specific enzyme corresponds to a terminal residue (i.e., exoprotease) or else to one of aminoacids within the sequence (i.e., endoprotease).
1.2. Endoproteases targeted for the development of anti-viral strategies
The activity of several endoproteases ensures viral infection, involving host and viral proteases which belong to the classes of serine- and cysteine-proteases, respectively. Both proteases of the host cell (which are supposed to assist the virus during the intracellular and extracellular phases of its cycle) and those of the virus act in a concerted fashion to regulate and coordinate specific steps of the viral propagation, such as (i) the entry and the replication of the virus, (ii) the maturation of the polyprotein and (iii) the assembly of the secreted virions for further diffusion [5] (Fig. 2 ).

Diagram of the involvement of host and viral proteases in SARS-CoV-2 life cycle. Activation of coronavirus spike proteins by host cell proteases occurs at different stages in the viral life cycle and in different cell localizations. The ACE2-dependent infectious entry at the cell membrane is triggered through the S protein cleavage by host proteases: furin (1) and/or TMPRSS2 (2). Intracellular activation of S protein is mediated by cathepsin in lysosomes (3) and/or by Furin in trans-Golgi network (TGN) (4). After the receptor recognition, the viral genome is released into the cytoplasm of the host cell (5), RNA attaches directly to the host ribosome for translation of two polyproteins (not shown). Polyprotein (pp) maturation into mature fragments is catalysed by viral Cys proteases (Mpro and PL pro) (6). RNA is translated into DNA and inside the nucleus (N) replication amplifies the number of virus genome copies (7). The viral genome produces pps, which help to take command over host ribosomes for their own translation process; protein biosynthesis starts at the endoplasmic reticulum (ER) and follows the constitutive secretory pathway along Golgi compartments (8). The virion assembly occurs (9) and the newly packed viral particles can egress (10).
The spike glycoproteins are responsible for the crown-like appearance of Coronavirus particles (Fig. 3 A), playing a crucial role for the entry of the viral genome inside the host cell (Fig. 2). The first critical step is the binding of the homotrimeric S protein with its specific cellular receptor, which triggers a cascade of proteolytic events leading to the fusion of cell and viral membranes. Similarity in structure and sequence with SARS-CoV and in vitro binding measurements indicate that SARS-CoV-2 S protein shows an improved binding for the receptor of angiotensin converting enzyme2 (ACE2), identifying it as the main host receptor [8]. The S protein is synthesized as an uncleaved precursor which includes two functionally distinct domains (i.e., S1 and S2 domains) that are responsible for receptor binding and triggering of the fusion event, respectively (Fig. 3B).

Open in a separate windowFig. 3
(A): Schematic representation of coronavirus particle. Spike proteins are highly glycosylated type I transmembrane protein, which assemble into trimers on the virion surface to form the distinctive “corona” (crown-like) appearance. (B): Domain organization and cleavage sites of the coronavirus Spike monomer (S). The ectodomain of all CoV spike proteins share the same organization in two domains, that is a N-terminal domain, named S1 and responsible for receptor binding, and a C-terminal S2 domain responsible for fusion. The domain organization of the S monomer consists of a signal peptide (SP), the N-terminal domain (NTD), the receptor-binding domain (RBD), the fusion peptide (FP), the internal fusion peptide (IFP), the heptad repeat 1/2 (HR1/2), and the transmembrane domain (TM).The region between the two domain is termed S1/S2 site. (C): Sequence of S1/S2 cleavage site of S protein from SARS-CoV-2. The four amino acid insertion (SPRRs), unique to SARS-CoV-2, is marked in yellow, the conserved S1/S2 cleavage site is marked in grey. (D): Comparative sequences of S protein cleavage sites. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The inactive CoV S protein acquires both cellular receptor binding and fusion function upon cleavage events at different sites, which can be carried out by multiple proteases at multiple sites in different cell compartments [10], [9] (see Fig. 2). Importantly, depending on CoV strain and cell type, CoV S protein is activated at a specific cell localization by one or several host proteases, including furin, trypsin, cathepsin L, transmembrane protease serine protease2 (TMPRSS2), TMPRSS4, or human airway trypsin-like protease (HAT) [11] (see Fig. 2). Exploiting redundant pathways to activate surface glycoproteins, the activating cleavage is mediated by multiple host membrane proteases via two distinct pathways, namely either (i) the late endosomal pathway, using cathepsins, and/or (ii) the cell-surface or early endosome pathway, using transmembrane serine proteases (e.g., TMPRSS2 and pro-protein convertase furin) (Fig. 2).
It has been suggested that the surface route is preferred under natural conditions, while repeated passages in cultured cells in vitro appears to exert a selective pressure in favour of virions bearing a greater capacity to invade the target cell via late endosomes [12], [9], [13]. Thus, to activate the fusion machinery of the viral S protein the cooperation in space and in time of multiple membrane proteases is demanded; the actually involved pool depends on both the virus strain and the specific host cell type expression profile of proteases (thus changing for each cell type).
Among host proteases, involved in the viral infection, furin is the one most widely present, being constitutively expressed in a variety of cell types. It cycles from the trans Golgi network (TGN)/endosomal compartments and cell surface, and it is known to accumulate in the TGN (where it is supposed to fulfil its proteolytic activity) [14](Fig. 2). Nevertheless, recently it has been also detected linked to the membrane of oral and airway epithelial cells [16], [15].
Unlike close relatives, SARS-CoV-2 can promptly infect a broad spectrum of human cell types, spanning from lung cells to endothelial, conjunctival and gut cells, with the respiratory district being the main target, displaying the peculiar ability to infect even the upper respiratory tract. The efficient spreading of virus relies on the protease arsenal of host cells which mediate the propagation of viral infection. The expression profile of furin and ACE2 in human cells could explain why SARS-CoV-2 is so efficient in spreading virus particles, since they are present throughout the body in endothelial cells with particularly increased levels in cells lying in alveoli and small intestine[17]. Moreover, SARS-CoV-2 S protein possesses a peculiar insertion of four amino acids (i.e., Ser-Pro-Arg -Arg-Ala-Arg689↓, see [18] and Fig. 3C), which has been identified as an additional cleavage site for the specificity of furin activity, strengthening the idea that this enzyme plays a dominant relevance in SARS-CoV-2 viral infection [13], [19], [20], [21].
Therefore, furin may play either (a) a role in the first entry of the virus, thanks to its topological location at the outer membrane (which would allow the formation of the ternary complex with ACE2, i.e., furin:SARS-CoV-2S:ACE2), and/or (b) during the transport of virions along the secretory pathway, further facilitating the virus diffusion (Fig. 2). This co-expression has been detected in airway epithelia, cardiac tissues and enteric canals [16], envisaging the possibility that in these districts the role of furin in favouring the virus cell entry is relevant, providing a cellular and molecular basis for the comprehension of the major clinical effects of COVID-19 in the tissues where these cell types are located.
A key discovery in understanding the mechanism of SARS-CoV-2 infection concerns the role of the androgen-responsive transmembrane serine protease 2 (TMPRSS2), that is expressed by specific epithelial tissues (including those of the respiratory and digestive tracts), facilitating the SARS-CoV-2 entry in the human airways by cleaving the viral spike (S) protein [22], [23], [19] (Fig. 2).
Beside host proteolytic enzymes, two viral proteases, namely Mpro and PLpro (involved in the maturation of viral polyprotein) are also recognized as important drug target(s). In particular, Mpro has been found to play a prominent role in the viral gene expression and replication, thus becoming an attractive target for anti-CoV-2 drugs. Notably, its quaternary structure renders Mpro ideal for rational drug design strategies against SARS-CoV-2, as there is a correlation between homodimer formation and the enzyme catalytic activity. Each protomer contains an antiparallel β-barrel structure, which has a folding scaffold similar to other viral chymotrypsin-like proteases. However, unlike chymotrypsin, the active site of SARS-CoV-2 Mpro contains a catalytic cysteinyl residue instead of a serine residue.
It must be stressed that although the endoprotease classes show a variety of catalytic sites (see above) and distinct protein folding, functional similarity can be found across evolutionary distant species (from viruses to humans) [6], thus representing a caveat in the development of effective COVID-19 therapeutic strategies.
Further, structural and evolutionary analyses indicate that SARS-CoV-2 Mpro is a highly conserved viral protein, which recognizes the sequence Leu-Met-Phe-Gln↓Ser-Gly-Ala while no human proteases share the same specificity [24]. This unique feature makes Mpro an even more attractive target for a broad inhibition of multiple stages in the viral life cycle (such as viral formation, progression of the viral infection and reproduction of virions).
Overall, two very attractive processes (which indeed represent important targets for designing anti-viral drugs), will be discussed here: (a) the proteolytic activation of the S protein (by furin and TMPRRS2), impairing the entry of viral genetic material inside the host cell, and (b) the activity of viral proteases (in particular Mpro), impairing the formation of mature viral proteins, which are required for the progression of the viral infection and replication of viruses.
For More Information: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7501082/